The continuing effort to identify pain-related genes has yielded considerable success. While classic approaches had identified at least 50 genes whose expression changed in various pain models (3), recent introduction of DNA microarray methodologies finally allowed to assess wide transcriptional genomic changes in chronic pain. So far, reports describing the profiling of transcriptional changes occurring in DRG after axotomy (4-6) and spinal nerve ligation (7, 8) have been published. The consensus was that the abundance of galanin, vasoactive intestinal peptide and neuropeptide Y as well as other transcripts including RT1.Mb, complement C1q ß and peripheral-type benzodiazepine receptor was affected by the development of neuropathic pain. The changes observed in the expression of neuropeptides were in agreement with previous reports (9, 10) and up-regulation of immune response genes had previously been associated with the development of neuropathic pain as well (11). Furthermore, it was reported that the expression of µ- (4) as well as
Defining the differences between the gene-expression profiles associated with different types of chronic pain would allow for better understanding of etiology of neuropathic pain. Nevertheless, we found no reports on global gene expression profiles in chronic inflammatory pain. Previous attempts to directly compare gene expression profiles in neuropathic and inflammatory pain have been limited to immunohistochemical studies (12). Therefore, we have performed a microarray screening for genes with different expression in rat models of neuropathic pain (chronic constriction injury) and inflammatory pain (injection of complete Freund's adjuvant) in the lumbar section of the rat spinal cord.
Animals
Experiments were performed on male Wistar rats (350-400g) housed in groups of 6-8 animals in cages lined with sawdust bedding under a standard 12-h/12-h light/dark cycle (08:00-20:00 h) with food and water available ad libitum. All experiments were conducted during the light phase, between 8:00 and 13:00. All experimental works were performed according to the ethical standards of the Declaration of Helsinki and International Association for the Study of Pain (22), and were approved by the local bioethics committee.
Animal models of neuropathic and inflammatory pain
Chronic constriction injury (CCI) was produced by tying four ligatures around the sciatic nerve (23) under sodium pentobarbital anesthesia (60 mg/kg, i.p.). The biceps femoris and the gluteus superficialis were separated and the right sciatic nerve was exposed. The ligatures (4/0 silk) were tied loosely around the nerve with 1 mm spacing, until they elicited a brief twitch in the respective hind limb. The sciatic nerve ligation decreased paw withdrawal threshold to tactile stimulation (in grams) with von Frey filaments in all rats (day 3 - injured paw 7.15±2.34 vs. control paw 20.5±2.24; day 14 injured paw 2.1±0.54 vs. control paw 23.25±1.58). Time-course curve of development of CCI-induced allodynia was published elsewhere (24).
Inflammation was induced by injection of 0.15 ml of complete Freund's adjuvant (CFA, Calbiochem, Darmstadt, Germany) into the plantar surface of the right hind limb of rats under brief halothane anesthesia (2-3% (v/v), 5l per min) for 2-3 min in a Plexiglas chamber (25). The inflammation remained confined to the inoculated paw throughout the observation period. The injection of CFA decreased paw withdrawal threshold to mechanical stimulation (in grams) in paw-pressure test in all rats (day 3 - inflamed paw 114±0.27 vs. control paw 244±0.46; day 14 inflamed paw 204±0.75 vs. control paw 262±0.65).
Five groups of animals were used in this study. The rats with chronic constriction injury and injected with CFA were allowed to survive for 3 and 14 days (30 rats for each time point; at each time point twenty rats were used for array hybridization and ten for qPCR). A group of 30 naive animals were used as the reference group. No deaths occurred during the experimental period in any of the groups.
Microarray screening of gene expression
Animals were sacrificed either on the 3rd or 14th day after adjuvant injection or nerve ligation. Ipsilateral lumbar (L4-L6) dorsal part of the spinal cord was isolated as well as ipsilateral lumbar (L5-L6) dorsal root ganglions (DRG) were carefully excised. After extraction tissues were rapidly frozen on dry ice and stored at -70°C until mRNA extraction procedure. Total RNA was isolated by acid guanidinium thiocyanate/phenol/chloroform extraction (26). RNA quality was assessed by agarose gel electrophoresis. Total RNA was tested for genomic DNA contamination by means of PCR with primers spanning a gene fragment with short intron in its sequence. RNA concentration was quantified by measuring absorbance at 260 nm. In order to prepare the labeled probe for hybridization, total RNA samples from 9-12 animals (~50 µg) were pooled and poly(A) RNA was purified using the Atlas Pure Total RNA Labeling System (BD Biosciences Clontech, San Jose, CA, USA) with streptavidin-coated magnetic beads and biotinylated oligo (dT) according to the manufacturer's instructions. For each group two separate pools of total RNA were obtained and converted to cDNA. Each cDNA was hybridized to independent microarray replicate. Poly(A) RNA was subjected to reverse transcription reaction (42°C for 30 min, MMLV Reverse Transcriptase, BD Biosciences) in the presence of random hexamer primers and [alpha-33P] dATP 10 µCi/µl (Perkin Elmer, Boston, MA, USA) according to the BD Atlas Plastic Microarrays User Manual. Positive control for labeling and hybridization reactions was provided with random primer mix. The labeled probe was purified from excess probe using spin columns provided in the kit. Total radioactivity of the labeled probe varied between 40-80x106 c.p.m. The labeled probe was added to 15 ml of BD PlasticHyb Hybridization Solution and hybridized with the Atlas Rat 4k arrays (BD Biosciences) in roller bottles (#308-8, LAB-LINE) at 60°C for 16 h at 20 rpm in a LAB-LINE hybridization oven. Arrays were washed with 2×SSC (0.3 M NaCl, 0.03 M trisodium citrate), 0.1% (w/v) SDS twice, then with 0.1xSSC (0.015 M NaCl, 0.0015 M trisodium citrate), 0.1% (w/v) SDS and finally with 0.1xSSC (0.015 M NaCl, 0.0015 M trisodium citrate) according to the manual. Plastic arrays were dried and exposed to BAS-SR 2025 Imaging Plates (Fuji Photo Film Co, Tokyo, Japan) for 1 to 7 days depending on signal intensity. Hybridization signals were scanned with phosphor imager FUJI BAS-5000 with high resolution (25 µm) and 16 bit gray scale depth. DNA array scans were saved as tagged image file format (TIFF) and analyzed with ArrayVision 8.0 Rev4.0 (Imaging Research, Ontario, Canada). Raw intensities were processed with 'SNOMAD', a collection of algorithms for normalization and standardization of DNA microarray data (27). Briefly, local background was subtracted from spot intensities, and then all positive values were subjected to the complete SNOMAD procedure with default parameters. Only the background correction step was omitted in SNOMAD since it has already been performed during pre-processing.
We have found that the number of spots detected on arrays has varied considerably from one array experiment to another, especially in case of spots corresponding to lower abundance genes. Additionally, normalization of array results from different experimental sets is problematic. It leads to a situation where only spots that were detected in all experiments can be analyzed. Therefore, we have decided to employ a simple approach for selecting signals corresponding to transcripts with changed transcription. All arrays from a single set were normalized in relation to the result from naive animals and z-scores were calculated. Only those genes with more than twofold change were considered to show differential expression. The highest quality set was selected as reference, while the remaining set was used as validation. Hierarchical clustering was performed using dChip 1.3 (28).
Reverse transcription Real-Time PCR reactions
Total RNA from ipsilateral lumbar (L4-L6) dorsal part of the spinal cord of two animals was pooled and used as separate sample for qPCR experiment. In addition, gene expression was measured in the pooled ipsilateral lumbar (L5-L6) dorsal root ganglions (4 or 10 per sample). Reverse transcription Real-Time PCR reactions (qPCR) were performed using Applied Biosystems TaqMan method, with TaqMan Reverse Transcription Reagents and TaqMan PCR Universal Master Mix (Applied Biosystems, Foster, CA, USA). Reactions were run on a Real-Time PCR iCycler device (BioRad, Hercules, CA, USA) with the 3.0a software version. The following TaqMan Assay-on-Demand primers and probes were used: calcitonin/calcitonin-related polypeptide, alpha, Rn00569199_m1; ICAM-1, Rn00564227_m1; chemokine-like receptor 1, Rn00573616_s1; TIMP-1, Rn00587558_m1; peripherin 1, Rn00561807_m1 and ß-2 microglobulin, Rn00560865_m1 as control. For each reaction, cDNA synthesized on 250 ng of total RNA template was used. A dilution curve to assess reaction efficiency has been prepared for each assay, efficiency values ranged from 1.65 to 2.0. Threshold cycle values were calculated automatically with default parameters. The abundance of RNA was calculated as 2-(threshold cycle-1). qPCR data were analyzed using one-way ANOVA followed by Tukey post-test.
Screening for genes with different expression in the dorsal horn of the spinal cord in the neuropathic or inflammatory pain was performed using the BD Atlas Rat 4k microarrays. Each array contains oligonucleotide probes representing 4 thousand of known genes. The labeled cDNA was prepared from RNA extracted from the ipsilateral dorsal part of L4-L6 spinal cord section, and after hybridization the images of arrays were normalized as described in Materials and Methods. Array screening indicated that 41 probes had z-score values larger than 3.5 or lower than -3.5, either in comparison to naive or between neuropathic and inflammatory pain models. Hierarchical clustering of the selected probes (Fig. 1) branched into 4 main groups, (1) transcripts with decreased abundance in neuropathic pain, (2) increased expression in both types of pain, (3) with decreased abundance on the 3rd day of inflammatory pain model and (4) with increased abundance only in neuropathic pain. Four out of seven transcripts with an increased abundance exclusively in neuropathic pain (branch 4 in Fig. 1), RT1.Ma, RT1.Mb, tissue inhibitor of metalloproteinase 1 (TIMP-1) and C1qß are associated with immune response and microglia activation. Descriptions of functions most relevant to pain or nerve tissue damage for each transcript are listed in Table 1. Based on literature, genes were arbitrarily grouped in six categories: vesicle exocytosis (10 transcripts), activation of immune response and microglia (8 transcripts), cytoskeleton (5 transcripts), secreted peptides and peptide processing (4 transcripts), nerve tissue injury (5 transcripts) and others (8 transcripts).
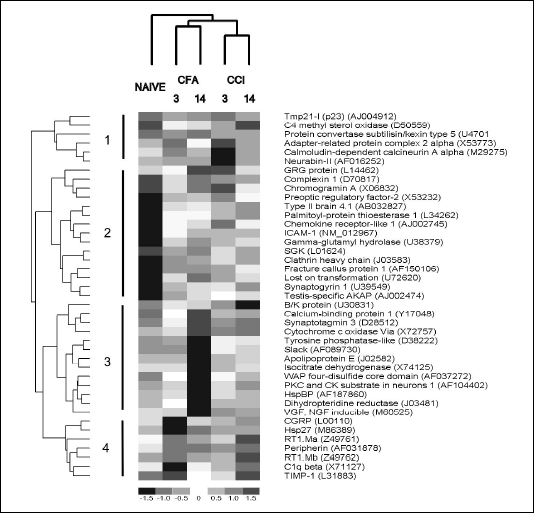 |
| Fig. 1. Hierarchical clustering of transcripts selected from expression profiling of neuropathic and inflammatory pain. The 41 genes with z-score between groups lower than -3.5 or greater than 3.5 were clustered with the dChip software based on Euclidean distances between expression patterns. Columns represent expression profiles in naive animals, rats after 3 or 14 days of complete Freund's adjuvant injection (CFA3 and CFA14) or chronic constriction nerve injury (CCI3 and CCI14 respectively). Each row corresponds to a single gene, with the colors of rectangles representing normalized expression on the scale shown below. Gene names with GeneBank accession numbers are listed to the right. Numbers and corresponding vertical lines mark the 4 main branches in the dendrogram from hierarchical clustering. |
| Table 1. Functional classification of genes selected from array analysis |
 |
Five genes, namely TIMP-1, ICAM-1, CGRP, chemokine-like receptor 1 and peripherin were selected for further validation with qPCR based on the largest average difference in expression between neuropathic and inflammatory pain.
The increased abundance of four transcripts, TIMP-1, ICAM-1, CGRP and chemokine-like receptor 1 in the dorsal section of the lumbar spinal cord after sciatic nerve ligation was confirmed by qPCR on samples independent from those used for microarray hybridization (Fig. 2). Expression of all four transcripts was significantly higher when compared to corresponding samples from animals with inflammatory pain, though not always versus naive, like in the case of ICAM-1. In all four cases, the measured abundances followed the same trend as indicated by microarrays (Fig. 1 and Fig. 2). Expression of peripherin was not found to be significantly changed, though there was a tendency towards a decrease in the inflammatory pain model on the 14th day (F(4,15)=3.185, P=0.0442, Tukey post-test N.S.).
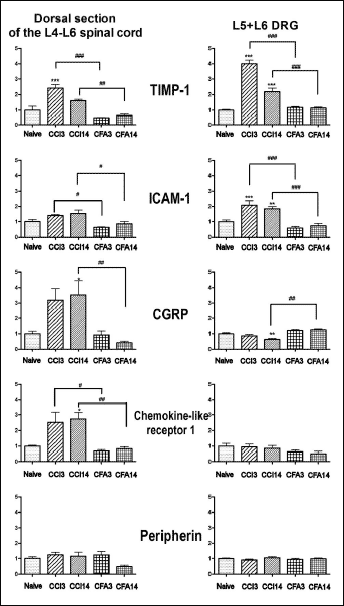 |
Fig. 2. Reverse transcription RealTime-PCR reactions (qPCR) analysis of expression of tissue inhibitor of metalloproteinase 1 (TIMP-1), intercellular adhesion molecule 1 (ICAM-1), calcitonin gene-related peptide (CGRP), chemokine-like receptor 1 gene and peripherin in the lumbar spinal cord and dorsal root ganglia (DRGs). The bars represent normalized averages derived from the threshold cycle in qPCR. CCI3 and CCI14 correspond to samples obtained on the 3rd and 14th day of chronic constriction injury, while CFA3 and CFA14 represent the 3rd and 14th day after injection of complete Freund's adjuvant. Data are expressed as the mean ± SEM. In the spinal cord, the average was calculated from 5-6 samples and each sample was prepared from two separate spinal cord fragments. In case of DRGs, there were 8-12 samples (4-10 of each L5 and L6). * P<0.05, **P<0.01, ***P<0.001 in comparison to naive group of animals (ANOVA, Tukey test), #P<0.05, ##P<0.01, ###P<0.001 indicate statistical significance between the corresponding groups in neuropathic and inflammatory pain (ANOVA, Tukey post-test). |
Additionally, the expression of the five selected genes was measured in the L5-L6 DRGs. Abundance of TIMP-1 and ICAM-1 transcripts was significantly increased (P<0.001) on the 3rd day of chronic constriction nerve injury both versus 3rd day of inflammatory pain model and naive rats (Fig. 2). In both cases, on the 14th day of neuropathic pain model mRNA, the abundances were still higher than on the 14th day of inflammatory pain model and in naive animals. A reverse trend was observed with CGRP in the L5-L6 DRGs. CGRP mRNA abundance was significantly lower (P<0.01) on 14th day of chronic constriction nerve injury as compared to the 14th day after injection of complete Freund's adjuvant and naive rats (Fig. 2). The amount of CGRP transcript in the DRGs was over tenfold greater than in the spinal cord, as estimated from threshold cycle values from qPCR. No differences in the expression of chemokine-like receptor 1 or peripherin genes were observed in the DRGs.
The presented array analysis of global gene expression indicates that both neuropathic and inflammatory pains are associated with a dramatic shift in the regulation of secretory vesicle trafficking in the spinal cord. At least 10 of the 41 genes with changed expression are directly involved in the processing of secretory vesicles, another two are proteases involved in processing of peptides and two correspond to the secreted peptides (Table 1). In most cases expression of those genes was higher in inflammatory and neuropathic pain in comparison to naive animals, with a notable exception of two genes from the synaptotagmin family (synaptotagmin 3 and B/K protein, Fig. 1). Nevertheless, the changes in expression of secretory vesicle trafficking-related genes followed no clear pattern differentiating neuropathic and inflammatory pain.
On the other hand, genes associated with immune response and activation of the microglia were up-regulated in the spinal cord almost exclusively after chronic constriction nerve injury and thus appear to be the markers of neuropathic pain. The four immune response- and microglia-associated genes (RT1.Ma, RT1.Mb, C1q ß and TIMP-1) that were classified as the fourth branch according to the clustering result (Fig. 1) had higher expression on the third day after operation, and there was a tendency to decrease on the 14th day. The observed up-regulation of the immune response genes is in agreement with earlier reports (8, 11). Previously Schafer et al. (13) reported that constitutive and the ischemia-induced C1q biosynthesis was restricted to brain microglia. It has been postulated that complement activation is a causative factor in neurodegeneration (14). On the other hand, the expression of the GRG protein, a member of the Groucho family which acts as a repressor of the transcription factor NF-
The expression pattern of the immune response- and microglia activation-related genes in the DRGs was similar. The abundance of TIMP-1 and ICAM-1 mRNAs was increased on the 3rd day of chronic constriction injury in the DRGs, and similarly to the spinal cord on 14th day after injury the levels of both transcripts were still elevated in comparison to naive animals. As for the chemokine-like receptor 1, its mRNA was barely detectable in the DRGs and no changes in expression were found (Fig. 2).
Differences in expression profiles between neuropathic and inflammatory pain were also found among the genes classified as cytoskeleton-related, and all appear to interact with actin filaments (Table 1). Four out of five (calcium-binding protein 1, type II brain 4.1 protein, hsp27 and peripherin) had higher transcription after chronic constriction nerve injury. These changes could be associated with the rearrangement of pain sensory pathways that take place during the development of neuropathic pain. An increase in expression of hsp27 and TIMP-1 after spinal cord injury was previously reported (16).
CGRP is expressed in the sensory afferent neurons (17); some authors suggest its important role in spinal and peripheral mechanisms of chronic pain (18-20). Particularly interesting are the alterations in the CGRP mRNA levels in both DRG as well as in the dorsal spinal cord observed in our study. A significant decrease in CGRP mRNA levels was observed in DRGs of neuropathic rats in accordance with the previous data (9), which may underlay a marked reduction of CGRP containing fibres in the ipsilateral superfical layers of the dorsal horn where the primarly afferent nerve endings (with cell bodies in DRG) are present.
Interestingly, we measured a significant, over threefold increase in the CGRP mRNA level in the dorsal spinal cord ipsilateral to the nerve injury although relative abundance of the message was lower than that in the DRG. In accordance with our observations a distinct CGRP mRNA-positive neurons were observed in lamina III in both the ipsilateral and contralateral dorsal horn seven days after unilateral rhizotomy (21). Thus the results indicate that spinal nerve injury enhance expression of CGRP in the deeper laminae of the dorsal horn neurons which may contribute to the mechanism of hyperalgesia and sensitization of WDR neurons in the spinal cord dorsal horn (8). It further suggests that CGRP may contribute not only to induction phase of neuropathic pain, but also to its maintenance phase. Increased expression of the CGRP gene in the spinal cord could be one of the factors responsible for the maintenance of neuropathic pain symptoms. In agreement with these suggestions Bennett and co-workers (18) have shown that CGRP (8-37), a truncated version of CGRP that binds as antagonist to the CGRP receptors, was effective in alleviating mechanical and thermal allodynia in a dose-dependent manner after its intrathecal administration in rats with spinal hemisection. Furthermore, our results indicate that the persistence of neuropathic pain is closely associated with the activation of microglia and add chromogranin A and the GRG protein to the list of genes involved in this process.
Acknowledgements: This research was supported partially by statutory funds from the Ministry of Scientific Research and Information Technology (Warszawa, Poland) and by EU grant No. QLRT-2001-02919. The contribution of Foundation for Polish Science (FNP) to I. Obara is greatly acknowledged.
- Woolf CJ, Mannion RJ. Neuropathic pain: aetiology, symptoms, mechanisms, and management. Lancet 1999; 353:1959-1964.
- Dellemijn P. Are opioids effective in relieving neuropathic pain? Pain 1999; 80:453-462.
- Mogil JS, McCarson KE. Identifying pain genes: bottom-up and top-down approaches. J Pain 2000; 1(3 Suppl):66-80.
- Xiao HS, Huang QH, Zhang FX et al. Identification of gene expression profile of dorsal root ganglion in the rat peripheral axotomy model of neuropathic pain. Proc Natl Acad Sci U S A 2002; 99:8360-8365.
- Costigan M, Befort K, Karchewski L et al. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci 2002; 3:16.
- Yang L, Zhang FX, Huang F et al. Peripheral nerve injury induces trans-synaptic modification of channels, receptors and signal pathways in rat dorsal spinal cord. Eur J Neurosci 2004; 19:871-883.
- Valder CR, Liu JJ, Song YH, Luo ZD. Coupling gene chip analyses and rat genetic variances in identifying potential target genes that may contribute to neuropathic allodynia development. J Neurochem 2003; 87:560-573.
- Okazaki Y, Furuno M, Kasukawa T et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002; 420:563-573.
- Noguchi K, De Leon M, Nahin RL, Senba E, Ruda MA. Quantification of axotomy-induced alteration of neuropeptide mRNAs in dorsal root ganglion neurons with special reference to neuropeptide Y mRNA and the effects of neonatal capsaicin treatment. J Neurosci Res 1993; 35:54-66.
- Nahin RL, Ren K, De Leon M, Ruda M. Primary sensory neurons exhibit altered gene expression in a rat model of neuropathic pain. Pain 1994; 58:95-108.
- Sweitzer SM, White KA, Dutta C, DeLeo JA. The differential role of spinal MHC class II and cellular adhesion molecules in peripheral inflammatory versus neuropathic pain in rodents. J Neuroimmunol 2002; 125:82-93.
- Honore P, Rogers SD, Schwei MJ et al. Murine models of inflammatory, neuropathic and cancer pain each generates a unique set of neurochemical changes in the spinal cord and sensory neurons. Neuroscience 2000; 98:585-598.
- Schafer MK, Schwaeble WJ, Post C et al. Complement C1q is dramatically up-regulated in brain microglia in response to transient global cerebral ischemia. J Immunol 2000; 164:5446-5452.
- McGeer PL, McGeer EG. The inflammatory response system of brain: implications for therapy of Alzheimer and other neurodegenerative diseases. Brain Res Brain Res Rev 1995; 21:195-218.
- Ciesielski-Treska J, Ulrich G, Chasserot-Golaz S et al. Mechanisms underlying neuronal death induced by chromogranin A-activated microglia. J Biol Chem 2001; 276:13113-13120.
- Tachibana T, Noguchi K, Ruda MA. Analysis of gene expression following spinal cord injury in rat using complementary DNA microarray. Neurosci Lett 2002; 327:133-137.
- Wiesenfeld-Hallin Z, Hokfelt T, Lundberg JM et al. Immunoreactive calcitonin gene-related peptide and substance P coexist in sensory neurons to the spinal cord and interact in spinal behavioral responses of the rat. Neurosci Lett 1984; 52:199-204.
- Bennett AD, Chastain KM, Hulsebosch CE. Alleviation of mechanical and thermal allodynia by CGRP(8-37) in a rodent model of chronic central pain. Pain 2000; 86:163-175.
- Jang JH, Nam TS, Paik KS, Leem JW. Involvement of peripherally released substance P and calcitonin gene-related peptide in mediating mechanical hyperalgesia in a traumatic neuropathy model of the rat. Neurosci Lett 2004; 360:129-132.
- Ma W, Chabot JG, Powell KJ, Jhamandas K, Dickerson IM, Quirion R. Localization and modulation of calcitonin gene-related peptide-receptor component protein-immunoreactive cells in the rat central and peripheral nervous systems. Neuroscience 2003; 120:677-694.
- Tie-Jun SS, Xu Z, Hokfelt T. The expression of calcitonin gene-related peptide in dorsal horn neurons of the mouse lumbar spinal cord. Neuroreport 2001; 12:739-743.
- Zimmermann M. Ethical guidelines for investigations of experimental pain in conscious animals. Pain 1983; 16:109-110.
- Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain 1988; 33:87-107.
- Starowicz K, Bilecki W, Sieja A, Przewlocka B, Przewlocki R. Melanocortin 4 receptor is expressed in the dorsal root ganglions and down-regulated in neuropathic rats. Neurosci Lett 2004; 358:79-82.
- Stein C, Millan MJ, Herz A. Unilateral inflammation of the hindpaw in rats as a model of prolonged noxious stimulation: alterations in behavior and nociceptive thresholds. Pharmacol Biochem Behav 1988; 31:455-451.
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162:156-159.
- Colantuoni C, Henry G, Zeger S, Pevsner J. SNOMAD (Standardization and NOrmalization of MicroArray Data): web-accessible gene expression data analysis. Bioinformatics 2002; 18:1540-1541.
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A 2001; 98:31-36.
- Horer J, Blum R, Feick P, Nastainczyk W, Schulz I. A comparative study of rat and human Tmp21 (p23) reveals the pseudogene-like features of human Tmp21-II. DNA Seq 1999; 10:121-126.
- Tucker KL, Nathanson K, Kirchhausen T. Sequence of the rat alpha c large chain of the clathrin associated protein complex AP-2. Nucleic Acids Res 1990; 18:5306.
- McMahon HT, Missler M, Li C, Sudhof TC. Complexins: cytosolic proteins that regulate SNAP receptor function. Cell 1995; 83:111-119.
- Ahtiainen L, Van Diggelen OP, Jalanko A, Kopra O. Palmitoyl protein thioesterase 1 is targeted to the axons in neurons. J Comp Neurol 2003; 455:368-377.
- Kirchhausen T, Harrison SC, Chow EP et al. Clathrin heavy chain: molecular cloning and complete primary structure. Proc Natl Acad Sci U S A 1987; 84:8805-8809.
- Stenius K, Janz R, Sudhof TC, Jahn R. Structure of synaptogyrin (p29) defines novel synaptic vesicle protein. J Cell Biol 1995; 131(6 Pt 2):1801-1809.
- Mizuta M, Inagaki N, Nemoto Y, Matsukura S, Takahashi M, Seino S. Synaptotagmin III is a novel isoform of rat synaptotagmin expressed in endocrine and neuronal cells. J Biol Chem 1994; 269:11675-11678.
- Kwon OJ, Gainer H, Wray S, Chin H. Identification of a novel protein containing two C2 domains selectively expressed in the rat brain and kidney. FEBS Lett 1996; 378:135-139.
- Qualmann B, Roos J, DiGregorio PJ, Kelly RB. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell 1999; 10:501-513.
- Nowak FV. Cloning of two hypothalamic cDNAs encoding tissue-specific transcripts in the preoptic area and testis. Mol Endocrinol 1990; 4:1205-1210.
- Schmidt CJ, Sladek TE. A rat homolog of the Drosophila enhancer of split (groucho) locus lacking WD-40 repeats. J Biol Chem 1993; 268:25681-25686.
- Owman C, Lolait SJ, Santen S, Olde B. Molecular cloning and tissue distribution of cDNA encoding a novel chemoattractant-like receptor. Biochem Biophys Res Commun 1997; 241:390-394.
- Simmons D, Makgoba MW, Seed B. ICAM, an adhesion ligand of LFA-1, is homologous to the neural cell adhesion molecule NCAM. Nature 1988; 331:624-627.
- Alfonso C, Han JO, Williams GS, Karlsson L. The impact of H2-DM on humoral immune responses. J Immunol 2001; 167:6348-6355.
- Rivera S, Ogier C, Jourquin J et al. Gelatinase B and TIMP-1 are regulated in a cell- and time-dependent manner in association with neuronal death and glial reactivity after global forebrain ischemia. Eur J Neurosci 2002; 15:19-32.
- Gentleman RC, Carey VJ, Bates DM et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:R80.
- Seidenbecher CI, Langnaese K, Sanmarti-Vila L et al. Caldendrin, a novel neuronal calcium-binding protein confined to the somato-dendritic compartment. J Biol Chem 1998; 273:21324-21331.
- Yamakawa H, Ohara O. Comparison of mRNA and protein levels of four members of the protein 4.1 family: the type II brain 4.1/4.1B/KIAA0987 is the most predominant member of the protein 4.1 family in rat brain. Gene 2000; 248:137-145.
- Willis D, Li KW, Zheng JQ et al. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci 2005; 25:778-791.
- Thompson MA, Ziff EB. Structure of the gene encoding peripherin, an NGF-regulated neuronal-specific type III intermediate filament protein. Neuron 1989; 2:1043-1053.
- Oku R, Satoh M, Fujii N, Otaka A, Yajima H, Takagi H. Calcitonin gene-related peptide promotes mechanical nociception by potentiating release of substance P from the spinal dorsal horn in rats. Brain Res 1987; 403:350-354.
- Hawley RJ, Scheibe RJ, Wagner JA. NGF induces the expression of the VGF gene through a cAMP response element. J Neurosci 1992; 12:2573-2581.
- Cain BM, Vishnuvardhan D, Beinfeld MC. Neuronal cell lines expressing PC5, but not PC1 or PC2, process Pro-CCK into glycine-extended CCK 12 and 22. Peptides 2001; 22:1271-1277.
- Wang Y, Nimec Z, Ryan TJ, Dias JA, Galivan J. The properties of the secreted gamma-glutamyl hydrolases from H35 hepatoma cells. Biochim Biophys Acta 1993; 1164:227-235.
- Imaizumi K, Tsuda M, Wanaka A, Tohyama M, Takagi T. Differential expression of sgk mRNA, a member of the Ser/Thr protein kinase gene family, in rat brain after CNS injury. Brain Res Mol Brain Res 1994; 26:189-196.
- Hadjiargyrou M, Halsey MF, Ahrens W, Rightmire EP, McLeod KJ, Rubin CT. Cloning of a novel cDNA expressed during the early stages of fracture healing. Biochem Biophys Res Commun 1998; 249:879-884.
- Ciani E, Frenquelli M, Contestabile A. Developmental expression of the cell cycle and apoptosis controlling gene, Lot1, in the rat cerebellum and in cultures of cerebellar granule cells. Brain Res Dev Brain Res 2003; 142:193-202.
- Hayashi H, Campenot RB, Vance DE, Vance JE. Glial lipoproteins stimulate axon growth of central nervous system neurons in compartmented cultures. J Biol Chem 2004; 279:14009-14015.
- Uwabe K, Gahara Y, Yamada H, Miyake T, Kitamura T. Identification and characterization of a novel gene (neurorep 1) expressed in nerve cells and up-regulated after axotomy. Neuroscience 1997; 80:501-509.
- Turk R, t Hoen PA, Sterrenburg E et al. Gene expression variation between mouse inbred strains. BMC Genomics 2004; 5:57.
- Raynes DA, Guerriero V. Isolation and characterization of isoforms of HspBP1, inhibitors of Hsp70. Biochim Biophys Acta 2000; 1490:203-207.
- Larsen M, Ressler SJ, Lu B et al. Molecular cloning and expression of ps20 growth inhibitor. A novel WAP-type "four-disulfide core" domain protein expressed in smooth muscle. J Biol Chem 1998; 273:4574-4584.
- Ito A, Hashimoto T, Hirai M et al. The complete primary structure of calcineurin A, a calmodulin binding protein homologous with protein phosphatases 1 and 2A. Biochem Biophys Res Commun. 1989; 163:1492-1497.
- Joiner WJ, Tang MD, Wang LY et al. Formation of intermediate-conductance calcium-activated potassium channels by interaction of Slack and Slo subunits. Nat Neurosci 1998; 1:462-469.