Since a cancer test based on blood samples has become a major challenge of recent years, choosing the reliable and efficient method of plasma DNA quantification would be an essential step prior to any clinical evaluation of cell-free DNA measurement in cancer patients. As time-consuming radioimmunoassay (P32) and common bench UV-spectrophotometric method cannot detect DNA below nanogram levels, an ultrasensitive quantitative real-time PCR (qPCR) technique might be regarded as a standard method currently available for DNA quantification. It is characterized by high accuracy, reproducibility, and time effectiveness, but requires specific automated systems and remains expensive. Thus, simple DNA quantification using fluorescent dyes that selectively binds double-stranded DNA may prove an advantageous alternative in the context of cost and time effectiveness and procedure simplicity. Among the other commercially available fluorochromes, PicoGreen assay demonstrates exceptionally high detection limit up to 25 pg/ml and perfect linearity up to 1000 ng/ml, making the test appropriate for plasma/serum DNA analysis. The present study was designed to compare the performance of plasma DNA quantification by the use of the PicoGreen assay and the real-time PCR, representing approaches of total vs. amplifiable DNA measurement, respectively.
The study was performed in accordance with the guidelines of the Declaration of Helsinki for Human Research and was approved by a local Ethics Committee.
Collection of plasma samples and DNA extraction
Blood samples were drawn from 10 patients with resectable non-small cell lung cancer (NSCLC): (i) before the start of therapy, (ii) after neoadjuvant chemotherapy, and (iii) after surgery. Blood was collected in 10-ml EDTA tubes and processed within 1 h of collection by centrifugation at 1000 x g for 10 min in 4°C. Plasma was then separated and banked at -80°C in 0.5 ml aliquots. DNA was extracted from 2 ml plasma samples with QIAmp DNA Blood Midi kit (Qiagen, Germany) according to the manufacturer’s instructions and stored at -20°C.
Quantification of plasma total DNA by direct fluorescent PicoGreen staining
DNA quantification was performed using PicoGreen dsDNA kit (Molecular Probes, U.S.A.), according to the manufacturer’s instructions. Briefly, PicoGreen dye was diluted 1:200 with TE (pH=7). Each reaction contained 50 µl of a dye solution plus a sample DNA made up to 50 µl in TE. Each sample DNA was analyzed in two duplicated dilution series. Standard curves were constructed by serial dilution of lambda DNA stock provided by the manufacturer. Black microtiter plates (Nunc, Denmark) were read in FLx800 fluorometer (Bio-Tek, USA) at an emission wavelength of 520 nm and excitation of 480 nm. Blank values were subtracted and replicates averaged for each sample.
Quantification of plasma amplifiable DNA by real-time PCR
The extracted plasma DNA was measured quantitatively by real-time qPCR in a SYBR Green I and TaqMan probe detection approach using human ß-actin gene as the amplifying (99 bp) target. The primer and probe sequences were as follows: forward primer, 5’-CCA CAC TGT GCC CAT CTA CG-3’; reverse primer, 5’-AGG ATC TTC ATG AGG TAG TCA GTC AG-3’; probe, 5’-TET-ATG CCC TCC CCC ATG CCA TCC TGC GT-TAMRA-3’ (2). qPCR was performed with a Chromo4 Multicolor Real-Time PCR Detection System (Bio-Rad Laboratories, CA) and DNA samples were quantified from genomic DNA standard curves. QPCR reaction components for SYBR Green detection approach were as follows: 12.5 µl of iQ SYBR Green Supermix (Bio-Rad, Laboratories), 500 nM each primer, and 10 µl of extracted DNA. QPCR reaction components for TaqMan probe detection approach were as follows: 12.5 µl iQ Supermix (Bio-Rad Laboratories), 400 nM each primer, 200 nM probe, and 10 µl of extracted DNA. PCR cycling conditions for both detection approaches were as follows: 95°C for 12 min, followed by 95°C for 15 s and 64°C for 1 min, repeated for 40 cycles. All samples were processed in duplicate, and the mean value was used for quantification.
Statistical analysis
Statistical data evaluation was done with Statsoft-2005 and Statistica PL version 7. ANOVA was used to test statistical significance of differences and a post hoc Tukey test was used for multiple comparisons (P<0.05). Correlations between PicoGreen and real-time PCR quantification results were assessed using a non-parametric linear regression with no assumption about the distribution of the values (Spearman rank test).
PicoGreen assay performance
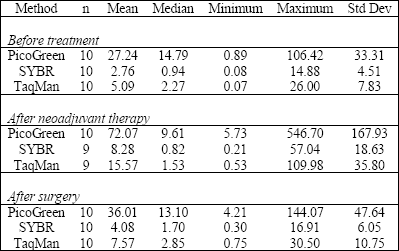
A total of 30 extracted plasma DNA samples were evaluated quantitively by direct PicoGreen staining in a microplate assay. The values of DNA concentration in plasma ranged from 0.89 up to 546.70 ng/ml (Table 1). The fit of lambda DNA standard curve was r2=0.998. The precision of test performance measured as a mean coefficient of variance (CV) was 3.61%.
| Table 1. Descriptive statistics for the data of plasma DNA concentration (ng DNA per 1 ml plasma) analysis by the use of PicoGreen and QPCR methods in relation to blood sample collection period. |
 |
Real-time PCR performance
Apart from the PicoGreen evaluation, concentration of DNA in plasma samples was measured by a real-time PCR method, using b-actin gene as the amplifying target. The values of DNA concentration in plasma ranged from 0.8 to 57.0 ng/ml by SYBR Green assay and from 0.7 to 110 ng/ml by TaqMan probe assay. One plasma DNA sample from a patient after neoadjuvant chemotherapy failed to give a PCR product in both SYBR Green and TaqMan approaches. Linearity of product amplification in qPCR assessed as the slope and correlation coefficient (r2) of the standard curve were -3.20, r2=0.999 and -3.50, r2=0.999 for the SYBR Green and TaqMan probe assays, respectively. The mean CV of threshold cycle (CVCT) for the qPCR method was 0.44% (SYBR Green) and 0.46% (TaqMan probe).
Plasma DNA evaluation
The mean levels of DNA content in plasma of 10 patients with NSCLC differed in relation to the phase of disease in which blood samples were collected (Table 1). The mean plasma DNA concentrations for cancer patients before treatment, after neoadjuvant therapy, and post-surgically were as follows: 27.2, 72.1, and 36.0 ng/ml by PicoGreen assay; 2.8, 8.3, and 4.1 ng/ml by SYBR Green-qPCR assay, and 5.1, 15.6, and 7.6 ng/ml by TaqMan probe-qPCR assay (Table 1). Seven of the 10 patients showed significant (P<0.05) differences in the plasma DNA content with respect to the phase of treatment (data not shown). The total DNA plasma content determined by PicoGreen proved to be several-fold higher than amplifiable DNA amount measured by qPCR (statistically significant for SYBR Green approach, P<0.03).
Correlation between PicoGreen and qPCR methods
Plasma DNA concentration values determined by PicoGreen assay correlated with real-time PCR quantification results with respect to DNA detection approach used. The PicoGreen method showed a high level of correlation with both SYBR Green (r=0.87, P<0.0001, Fig. 1A) and TaqMan probe approaches (r=0.94, P<0.0001; Fig. 1B). Additionally, the two DNA detection approaches of qPCR were correlated with each other (r=0.96, P<0.0001; Fig. 1C).
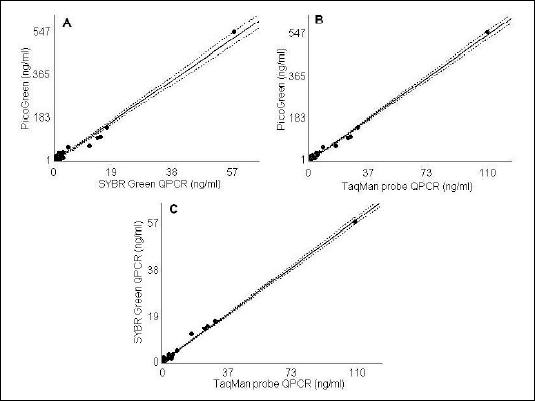 |
| Fig. 1. Spearman rank correlation graphs for the two plasma DNA concentration (ng/ml) quantification techniques. A - ß-actin qPCR with SYBR Green detection approach compared to PicoGreen assay (r=0.87, P<0.0001); B - ß-actin qPCR with TaqMan probe detection approach compared to PicoGreen assay (r=0.94, P<0.0001); and C - TaqMan probe compared to SYBR Green detection approach of ß-actin QPCR (r=0.96, P<0.0001). The 95% confidence intervals are shown for all correlation curves. |
In the last decade, numerous reports demonstrated that the quantification of free-circulating DNA in plasma/serum samples might be a promising biomarker in a number of pathologies (3). The occurrence of free DNA circulating in the blood of patients with lung cancer is one of the most intensively studied issues, due to a permanently high morbidity and mortality of the disease worldwide. Moreover, lung cancer stands out of a variety of benign and malignant diseases, because of its exceptionally high levels of free-circulating DNA detectable in plasma/serum samples (4). Since a diagnostic cancer test based on a molecular analysis of blood samples is appealing, the development and standardization of highly sensitive and cost-effective molecular biology techniques for plasma/serum DNA analysis is in demand.
In present study, free-circulating DNA concentration was measured in plasma samples by the use of two distinct techniques: direct fluorescent dye staining and real-time PCR. The plasma DNA samples were collected from NSCLC patients before the start of treatment, after neoadjuvant therapy, and post-surgically, and were expected to give a high scatter of result (as evidenced by the standard deviation values, Table 1). DNA extracted from plasma has been analyzed with PicoGreen, although the measurement of plasma DNA in situ, without extraction, has been reported by other authors (5). It has been demonstrated, however, that sensitive detection of plasma/serum DNA by PicoGreen method might be impaired by proteins causing high background fluorescence (6).
Our quantitative results are concordant with the data reported by other authors who used the PicoGreen (7) and qPCR (2) methods for evaluation of DNA concentration in plasma of patients with resectable NSCLC. Interestingly, the plasma DNA content determined by PicoGreen proved to be several-fold higher than the plasma DNA amount measured by qPCR. Such difference may be accounted for by the fact that the first method can detect nearly all DNA fragments (8), whereas the latter measures only amplifiable DNA. Similarly, Sozzi et al (9) evaluated circulating plasma DNA in lung cancer patients and reported the mean DNA concentration of 318 ng/ml using a colorimetric assay, but only 24 ng/ml, when quantitative real-time PCR of the human telomerase reverse transcriptase (hTERT) gene has been applied (10).
Importantly, in our study the PicoGreen method showed a very strong correlation with both SYBR Green (r=0.87, P<0.0001) and TaqMan probe (r=0.94, P<0.0001) qPCR assays (Fig. 1 A, B). Recently, in a study of Chiminqgi et al (11), PicoGreen method also proved to be highly correlated with qPCR of b-globin (r=0.81, P<0.0001) and cyclophilin gene (r=0.915, P<0.0001) as amplifying targets, and was capable to discriminate between the cancer patients (n=180) and healthy controls (n=58) according to their plasma DNA levels (P<0.0001). The lack of statistical significance between the quantitative results representing three clinical phases of NSCLC observed in the present study likely arose from a small patients group.
In summary, the quantitative real-time PCR method is characterized by high accuracy, reproducibility, and time-effectiveness. Consequently, it is regarded as a standard method currently available for DNA quantification. Nevertheless, the use of gene-specific primer or probe sequences requires the optimization of PCR reaction conditions. Also, the automated performance of the PCR setup is recommended due to the high sensitivity of the method, which makes it an expensive technique. The diversity of protocols, reagents, and real-time PCR devices in use hampers the comparison of data from different laboratories (3). PicoGreen dye has exceptionally high fluorescence enhancement when bound to dsDNA, gives a minimum background, and allows longer exposure times and assay flexibility due to good stability to photobleaching (8). Interestingly, the measurement of dsDNA stained by fluorescent PicoGreen dye using real-time PCR apparatus has been described (12). Contrary to qPCR, the use of intercalating fluorochromes for direct dsDNA quantification may be a rapid, effective, and inexpensive alternative that extremely well correlates with real-time PCR. The simple, uniform protocol and microplate assay format makes PicoGreen a convenient tool for a high throughput quantitative plasma/serum DNA assessment and a good starting point toward multicenter standardization in cancer plasma/serum DNA studies.
Acknowledgments: This study was supported by grant N401 174 31/3840 by the Polish Ministry of Science and Informatics.
Conflicts of interest: No conflicts of interest were declared regarding this article.
- Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids-a promising, non-invasive tool for early detection of several human diseases. FEBS Lett 2007; 581: 795-799.
- Herrera LJ, Raja S, Gooding WE et al. Quantitative analysis of circulating plasma DNA as a tumor marker in thoracic malignancies. Clin Chem 2005; 51: 113-118.
- Chorostowska-Wynimko J, Szpechcinski A. The impact of genetic markers on the diagnosis of lung cancer: a current perspective. J Thorac Oncol 2007; 2: 1044-1051.
- Holdenrieder S, Stieber P, Bodenmüller H et al. Nucleosomes in serum of patients with benign and malignant diseases. Int J Cancer 2001; 95: 114-120.
- Jiang N, Pisetsky DS. The effect of inflammation on the generation of plasma DNA from dead and dying cells in the peritoneum. J Leukoc Biol 2005; 77: 296-302.
- Chen JA, Meister S, Urbonaviciute V et al. Sensitive detection of plasma/serum DNA in patients with systemic lupus erythrematous. Autoimmunity 2007; 40: 307-310.
- Xie GS, Hou AR, Li LY, Gao YN, Cheng SJ. Quantification of plasma DNA as a screening tool for lung cancer. Chin Med J 2004; 117: 1485-1488.
- Ahn SJ, Costa J, Emanuel JR. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res 1996; 24: 2623-2625.
- Sozzi G, Conte D, Mariani L et al. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res 2001; 61: 4675-4678.
- Sozzi G, Conte D, Leon M et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003; 21: 3902-3908.
- Chiminqgi M, Moutereau S, Pernet P et al. Specific real-time PCR vs. fluorescent dyes for serum free DNA quantification. Clin Chem Lab Med 2007; 45: 993-995.
- Blotta I, Prestinaci F, Mirante S, Cantafora A. Quantitative assay of total dsDNA with PicoGreen reagent and real-time fluorescent detection. Ann Ist Super Sanita 2005; 41: 119-123.